Assessment of Biofilm Formation and Hypermucoviscosity in Klebsiella oxytoca from Clinical and Community Sources
DOI:
https://doi.org/10.32007/jfacmedbaghdad2454Keywords:
Biofilm, Community-acquired infections, Klebsiella oxytoca, String testAbstract
Background: Klebsiella oxytoca is an emerging pathogen causing hospital- and community-acquired infections. Hypermucoviscous phenotype and biofilm development are considered markers of hypervirulence, causing invasive infections in healthy adults
Objective: The current study aimed to evaluate the ability of K. oxytoca isolates to form biofilms and incorporate hypermucoviscous characteristics.
Method: One hundred K. oxytoca isolates were collected from hospitals in the Medical City Teaching Hospital Complex, in Baghdad, Iraq. The isolates were identified by conducting manual and biochemical tests. The hypermucoviscous characteristic was analyzed by using the string test. The Congo red agar method was used to detect biofilm formation, while quantitative methods like tube adherence and microtiter plates were used to measure its strength. Statistical analysis was performed by using the SPSS Statistical package (Version 26; SPSS, IBM) and Microsoft Office Excel (2010) for drawing the figures except for the receiver operating characteristic (ROC) curve.
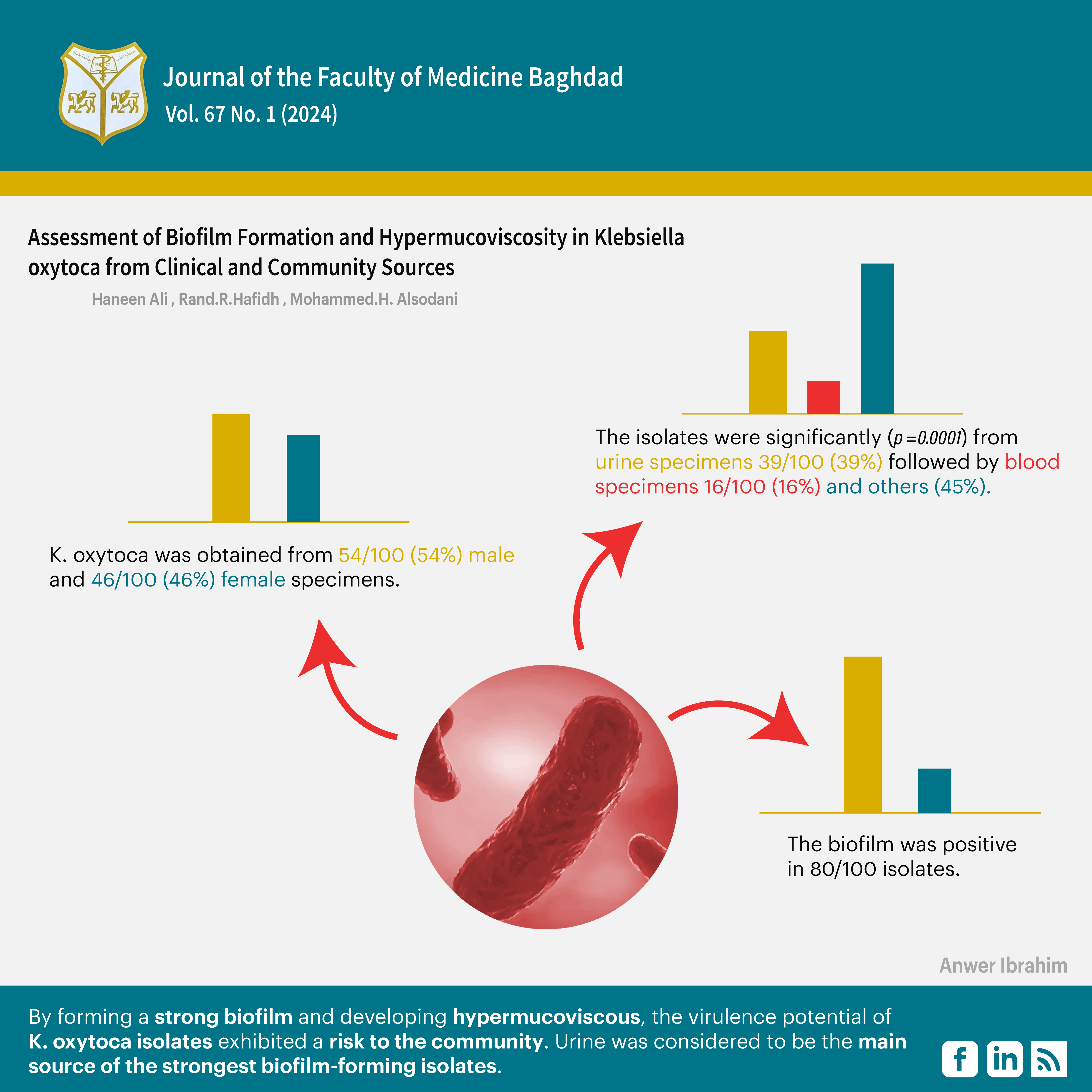
Results: K. oxytoca was obtained from 54/100 (54%) male and 46/100 (46%) female specimens. The specimens were from inpatients 62/100 (62%), and the rest, 38/100 (38%), were from outpatients. The isolates were significantly (P = 0.0001) from urine specimens 39/100 (39%) followed by blood specimens 16/100 (16%). The string test identified 14/100 (14%) isolates as hypermucoviscous with (21.1%) being from outpatients. The biofilm was positive in 80/100 isolates. By utilizing the tube method, a strong adhesion was detected in 23/80 (28.75%) isolates. The microtiter plate method (MTP) yielded a strong biofilm in 25/80 (31.25%) isolates. The strongest biofilm was found in isolates from urine specimens (40%).
Conclusion: By forming a strong biofilm and developing hypermucoviscous, the virulence potential of K. oxytoca isolates exhibited a risk to the community. Urine was considered to be the main source of the strongest biofilm-forming isolates.
Received: Aug., 2024
Revised: Dec., 2024
Accepted: Jan., 2025
Published: April 2025
Downloads
References
1. Janda JM, Abbott SL. The changing face of the family Enterobacteriaceae (Order:"Enterobacterales"): New members, taxonomic issues, geographic expansion, and new diseases and disease syndromes. Clinical microbiology reviews. 2021;34(2):10.1128/cmr. 00174-20. https://doi.org/10.1128/CMR.00174-20.
2. Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clinical microbiology reviews. 1998;11(4):589-603. https://doi.org/10.1128/CMR.11.4.589.
3. Neog N, Phukan U, Puzari M, Sharma M, Chetia P. Klebsiella oxytoca and emerging nosocomial infections. Current Microbiology. 2021;78(4):1115-23. https://doi.org/10.1007/s00284-021-02402-2.
4. Tomulescu C, Moscovici M, Lupescu I, Stoica RM, Vamanu A. A Review: Klebsiella pneumoniae, Klebsiella oxytoca. Rom Biotechnol Lett. 2021;26:2567-86. https://doi.org/10.25083/rbl/26.3/2567.2586.
5. Hussein RA, Al-Kubaisy SH, Al-Ouqaili MT. The influence of efflux pump, outer membrane permeability and β-lactamase production on the resistance profile of multi, extensively and pandrug resistant Klebsiella pneumoniae. Journal of Infection and Public Health. 2024 Nov 1;17(11):102544. https://doi.org/10.1016/j.jiph.2024.102544.
6. Ghasemian A, Mohabati Mobarez A, Najar Peerayeh S, Talebi Bezmin Abadi A, Khodaparast S, Mahmood SS. Expression of adhesin genes and biofilm formation among Klebsiella oxytoca clinical isolates from patients with antibiotic-associated haemorrhagic colitis. Journal of medical microbiology. 2019;68(7):978-85. https://doi.org/10.1099/jmm.0.000965.
7. Abebe G, Meseret G. 120-130 Detection of Biofilm Formation and Antibiotic Resistance in Klebsiella Oxytoca and Klebsiella Pneumoniae from Animal Origin Foods. International Journal of Microbiology and Biotechnology. 2020; 5:120-30. https://doi.org/10.11648/j.ijmb.20200503.17.
8. Li Y, Ni M. Regulation of biofilm formation in Klebsiella pneumoniae. Frontiers in Microbiology. 2023; 14:1238482. https://doi.org/10.3389/fmicb.2023.1238482.
9. Vestby LK, Grønseth T, Simm R, Nesse LL. Bacterial biofilm and its role in the pathogenesis of disease. Antibiotics. 2020;9(2):59. https://doi.org/10.3390/antibiotics9020059.
10. Wang G, Zhao G, Chao X, Xie L, Wang H. The characteristic of virulence, biofilm and antibiotic resistance of Klebsiella pneumoniae. International Journal of Environmental Research and Public Health. 2020;17(17):6278. https://doi.org/10.3390/ijerph17176278.
11. Yang S, Li X, Cang W, Mu D, Ji S, An Y, et al. Biofilm tolerance, resistance and infections increasing threat of public health. Microbial Cell. 2023;10(11):233. https://doi.org/10.15698/mic2023.11.807.
12. Silva NBS, Alves PGV, de Andrade Marques L, Silva SF, de Oliveira Faria G, de Araújo LB, et al. Quantification of biofilm produced by clinical, environment and hands' isolates Klebsiella species using colorimetric and classical methods. Journal of Microbiological Methods. 2021;185:106231. https://doi.org/10.1016/j.mimet.2021.106231.
13. Yadav B, Mohanty S, Behera B. Occurrence and Genomic Characteristics of Hypervirulent Klebsiella pneumoniae in a Tertiary Care Hospital, Eastern India. Infection and Drug Resistance. 2023:2191-201. https://doi.org/10.2147/IDR.S405816.
14. Shanthini T, Ramesh N. Phenotypic detection of hypermucoviscous phenotype in colistin-resistant Klebsiella oxytoca from Tamil Nadu. Research Journal of Biotechnology. 2020;15(10):36-40. https://www.researchgate.net/publication/344690872_Phenotypic_detection_of_hypermucoviscous_phenotype_in_colistin-resistant_Klebsiella_oxytoca_from_Tamil_Nadu
15. Lila ASA, Rajab AA, Abdallah MH, Rizvi SMD, Moin A, Khafagy E-S, et al. Biofilm lifestyle in recurrent urinary tract infections. Life. 2023;13(1):148. https://doi.org/10.3390/life13010148.
16. Maldonado NC, Silva de Ruiz C, Cecilia M, Nader-Macias M. A simple technique to detect Klebsiella biofilm-forming-strains. Inhibitory potential of Lactobacillus fermentum CRL 1058 whole cells and products. Communicating current research and educational topics and Trends in Applied Microbiology. 2007:52-9. https://citeseerx.ist.psu.edu/document?repid=rep1&type=pdf&doi=eeb7f3f1ce96f2053a24e3d241f6f8131e976717
17. Damian M, Usein C-R, Palade A-M, Ceciu S, Cosman M. Molecular epidemiology and virulence characteristics of Klebsiella pneumoniae strains isolated from hospital-associated infections. The Open Epidemiology Journal. 2009;2(1). https://doi.org/10.2174/1874297100902010069.
18. Emad R, Hafidh RR, Zaman MZ. Identification of Klebsiella oxytoca by VITEK-2 System in Baghdad Hospitals. J Fac Med Baghdad. 2023;65(4). https://doi.org/10.32007/jfacmedbagdad.2154.
19. Catalano A, Iacopetta D, Ceramella J, Scumaci D, Giuzio F, Saturnino C, et al. Multidrug resistance (MDR): A widespread phenomenon in pharmacological therapies. Molecules. 2022;27(3):616. https://doi.org/10.3390/molecules27030616.
20. Jebril NMT. Evaluation of two fixation techniques for direct observation of biofilm formation of Bacillus subtilis in situ, on Congo red agar, using scanning electron microscopy. Veterinary World. 2020;13(6):1133. https://doi.org/10.14202/vetworld.2020.1133-1137.
21. Harika K, Shenoy VP, Narasimhaswamy N, Chawla K. Detection of biofilm production and its impact on antibiotic resistance profile of bacterial isolates from chronic wound infections. Journal of global infectious diseases. 2020;12(3):129-34. https://doi.org/10.4103/jgid.jgid_150_19.
22. Stepanović S, Vuković D, Hola V, Bonaventura GD, Djukić S, Ćirković I, et al. Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. Apmis. 2007;115(8):891-9. https://doi.org/10.1111/j.1600-0463.2007.apm_630.x.
23. Dan B, Dai H, Zhou D, Tong H, Zhu M. Relationship between drug resistance characteristics and biofilm formation in Klebsiella pneumoniae strains. Infection and Drug Resistance. 2023:985-98. https://doi.org/10.2147/IDR.S396609.
24. Li Y, Wu Y, Li D, Du L, Zhao L, Wang R, et al. Multicenter comparative genomic study of Klebsiella oxytoca complex reveals a highly antibiotic-resistant subspecies of Klebsiella michiganensis. Journal of Microbiology, Immunology and Infection. 2024;57(1):138-47. https://doi.org/10.1016/j.jmii.2023.10.014.
25. Karimi K, Zarei O, Sedighi P, Taheri M, Doosti-Irani A, Shokoohizadeh L. Investigation of antibiotic resistance and biofilm formation in clinical isolates of Klebsiella pneumoniae. International Journal of Microbiology. 2021;2021. https://doi.org/10.1155/2021/5573388.
26. Nirwati H, Sinanjung K, Fahrunissa F, Wijaya F, Napitupulu S, Hati VP, et al., editors. Biofilm formation and antibiotic resistance of Klebsiella pneumoniae isolated from clinical samples in a tertiary care hospital, Klaten, Indonesia. BMC proceedings; 2019: Springer. https://doi.org/10.1186/s12919-019-0176-7.
27. Alvarez S, Stinnett JA, Shell CG, Berk SL. Klebsiella oxytoca isolates in a general hospital. Infection Control & Hospital Epidemiology. 1985;6(8):310-3. https://doi.org/10.1017/S0195941700063165.
28. Rawy DK, El-Mokhtar MA, Hemida SK, Askora A, Yousef N. Isolation, characterization and identification of Klebsiella pneumoniae from assiut university hospital and sewage water in assiut governorate, Egypt. Assiut Univ J Botany Microbiol. 2020;49(2):60-76. https://doi.org/10.21608/aunj.2020.221181.
29. Dawood FK, Al-Hayanni HS, Sultan MA. Contamination of Agricultural Soils in Some Baghdad Areas with Antibiotics Resistant Pathogenic Fecal Bacteria. J Fac Med Baghdad. 2023;65(3):220-6. https://doi.org/10.32007/jfacmedbagdad.2104.
30. Walker K.A. and Miller V.L., , . The intersection of capsule gene expression, hypermucoviscosity and hypervirulence in Klebsiella pneumoniae. Current Opinion in Microbiology. (2020);54, 95-102. https://doi.org/10.1016/j.mib.2020.01.006.
31. Jesmin Akter JA, Chowdhury A, Al-Forkan M. Study on prevalence and antibiotic resistance pattern of Klebsiella isolated from clinical samples in south east region of Bangladesh. 2014.
32. Rajivgandhi GN, Alharbi NS, Kadaikunnan S, Khaled JM, Kanisha CC, Ramachandran G, et al. Identification of carbapenems resistant genes on biofilm forming K. pneumoniae from urinary tract infection. Saudi Journal of Biological Sciences. 2021;28(3):1750-6. https://doi.org/10.1016/j.sjbs.2020.12.016.
33. Abood SH, Ibrahim IA. Comparison of biofilm formation by different species of Klebsiella. Pak J Biotechnol. 2017;14(4):531-5.
34. Alkhudhairy MK, Alshadeedi SM, Mahmood SS, Al-Bustan SA, Ghasemian A. Comparison of adhesin genes expression among Klebsiella oxytoca ESBL-non-producers in planktonic and biofilm mode of growth, and imipenem sublethal exposure. Microbial pathogenesis. 2019;134:103558. https://doi.org/10.1016/j.micpath.2019.103558.
35. Ahmed NAAST, Almohaidi AM. Investigation of biofilm formation ability and Assessment of cupB and rhlR Gene Expression in Clinical Isolates of Pseudomonas aeruginosa. Iraqi journal of biotechnology. 2022;21(2). View of Investigation of biofilm formation ability and Assessment of cupB and rhlR Gene Expression in Clinical Isolates of Pseudomonas aeruginosa
36. Ragupathi NKD, Sethuvel DPM, Dwarakanathan HT, Murugan D, Umashankar Y, Monk PN, et al. The influence of biofilm formation on carbapenem resistance in clinical Klebsiella pneumoniae infections: phenotype vs genome-wide analysis. BioRxiv. 2020:2020.07. 03.186130. https://doi.org/10.1101/2020.07.03.186130.
37. Gunardi WD, Karuniawati A, Umbas R, Bardosono S, Lydia A, Soebandrio A, et al. Biofilm-Producing Bacteria and Risk Factors (Gender and Duration of Catheterization) Characterized as Catheter-Associated Biofilm Formation. Int J Microbiol. 2021;2021:8869275. https://doi.org/10.1155/2021/8869275.
38. Emad R, Hafidh RR. Hypervirulent and the Multi-Drug Resistant Klebsiella oxytoca: A New Emerging Pathogen in Baghdad Hospitals, Iraq. Al-Anbar Medical Journal. 2024;20(1). https://doi.org/10.33091/amj.2024.143968.1409.
39. Mahato S, Mandal P, Mahato A. Biofilm Production by Uropathogens like Klebsiella spp and Pseudomonas spp and their Antibiotic Susceptibility. Birat Journal of Health Sciences. 2020;5(1):902-6. https://doi.org/10.3126/bjhs.v5i1.29609.
40. Khaertynov KS, Anokhin VA, Rizvanov AA, Davidyuk YN, Semyenova DR, Lubin SA, et al. Virulence Factors and Antibiotic Resistance of Klebsiella pneumoniae Strains Isolated From Neonates With Sepsis. Front Med (Lausanne). 2018;5:225. https://doi.org/10.3389/fmed.2018.00225.
41. Kong Q, Beanan JM, Olson R, Macdonald U, Shon AS, Metzger DJ, et al. Biofilm formed by a hypervirulent (hypermucoviscous) variant of Klebsiella pneumoniae does not enhance serum resistance or survival in an in vivo abscess model. Virulence. 2012;3(3):309-18. https://doi.org/10.4161/viru.20383.
42. Assefa M, Amare A. Biofilm-associated multi-drug resistance in hospital-acquired infections: A review. Infection and drug resistance. 2022:5061-8. https://doi.org/10.2147/IDR.S379502.
43. Khan J, Tarar SM, Gul I, Nawaz U, Arshad M. Challenges of antibiotic resistance biofilms and potential combating strategies: a review. 3 Biotech. 2021;11:1-15. https://doi.org/10.1007/s13205-021-02707-w.
Downloads
Published
Issue
Section
License
Copyright (c) 2025 Haneen A. Mustaf, Rand R. Hafidh, Mohammed H. Alsodani

This work is licensed under a Creative Commons Attribution 4.0 International License.











 Creative Commons Attribution 4.0 International license..
Creative Commons Attribution 4.0 International license..


