Molecular Detection of the mecA and some Virulence Determinants in Methicillin-Resistant Staphylococcus aureus
DOI:
https://doi.org/10.32007/jfacmedbagdad.6622282Keywords:
Methicillin-resistant Staphylococcus aureus, icaA gene, Coa gene, Hla gene, mecA geneAbstract
Background: Methicillin Resistant Staphylococcus aureus (MRSA) is globally acknowledged as a prominent contributor to both hospital-acquired and community infections. Understanding key virulence factors including coagulase production, hemolysis ability and biofilm formation, is crucial.
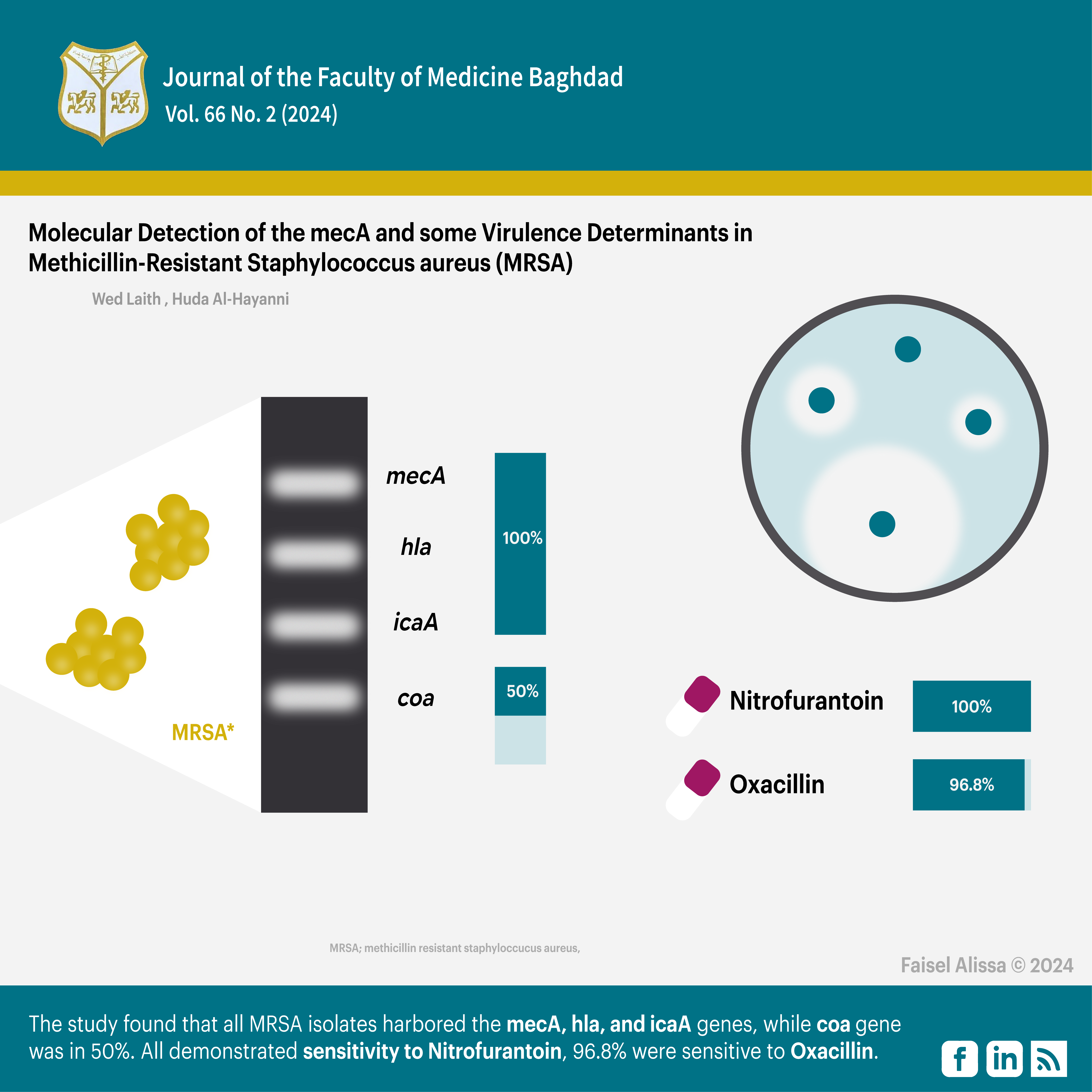
Objective: The study aimed to establish a molecular characterisation of mecA gene and virulence factors genes (hla, icaA, and coa) in clinical isolates of MRSA obtained from two hospitals in Baghdad.
Methods: A hundred and five isolates were obtained from clinical sources from November 2022 to March 2023 and their antibiotic sensitivity was assessed using the agar diffusion test against seven different antibiotics (Azithromycin, Ciprofloxacin, Nitrofurantoin, Rifampin, Trimethoprim, Ofloxacin and Oxacillin), Through Conventional Polymerase Chain Reaction, the presence of virulence factor genes including mecA, hla, icaA, and coa, was determined in MRSA isolates.
Results: All MRSA isolates (100%) harboured the mecA, hla, and icaA genes while the coa gene was recognized in 50% of the isolates. Regarding antibiotic susceptibility, all MRSA isolates (100%) demonstrated sensitivity to Nitrofurantoin. Additionally, 96.8% of the isolates were sensitive to Oxacillin.
Conclusion: Molecular detection of methicillin resistance genes and virulence genes can be used to diagnose MRSA isolates in hospitals. The presence of these genes may affect their pattern of sensitivity to antibiotics.
Received: Dec, 2023
Revised: Jan. 2024
Accepted: Feb. 2024
Published: July 2024
Downloads
References
1. Jenul C, Horswill AR. Regulation of Staphylococcus aureus Virulence. Microbiol Spectr 2019; 7(2). https://doi.org/10.1128/microbiolspec.gpp3-0031-2018.
2. Cheung GYC, Bae JS, Otto M. Pathogenicity and virulence of Staphylococcus aureus. Virulence 2021; 12(1): 547-569 https://doi.org/10.1080/21505594.2021.1878688.
3. Lynch JP, Zhanel GG. Escalation of antimicrobial resistance among MRSA part 1: focus on global spread. Expert Rev Anti Infect Ther 2023;21(2):99-113 https://doi.org/10.1080/14787210.2023.2154653.
4. Al-Hasnawi EA. Isolation of Staphylococcus aureusfrom ear swab in Iraqi children as a causative agent of Otitis externa. J Fac Med Baghdad 2017;59(3):258-61 https://doi.org/10.32007/jfacmedbagdad.593100.
5. Cetik Yildiz S. Staphylococcus aureus and Methicillin Resistant Staphylococcus aureus (MRSA) Carriage and Infections. Infectious Diseases. IntechOpen. 2023. http://dx.doi.org/10.5772/intechopen.107138http://dx.doi.org/10.5772/intechopen.107138.
6. Yamaguchi T, Nakamura I, Sato T, Ono D, Sato A, Sonoda S, et al. Changes in the Genotypic Characteristics of Community-Acquired Methicillin-Resistant Staphylococcus aureus Collected in 244 Medical Facilities in Japan between 2010 and 2018: a Nationwide Surveillance. Microbiol Spectr 2022;10(4): e02272-2. https://doi.org/10.1128/spectrum.02272-21.
7. Hami IA, Ibrahim KS. Incidence of Methicillin-Resistant Staphylococcus aureus (MRSA) Recovered from Patients with Urinary Tract Infections in Zakho City/ Kurdistan-Iraq. SJUOZ 2023;11(1), 91–97 https://doi.org/10.25271/sjuoz.2023.11.1.1041.
8. Hameed RM, Alafloogee JF, Ma’an GK. Bacteriological profile and antibiotics used for septic patients in Karbala, Iraq. Med J Babylon. 2021; 18(3):195-9. https://doi.org/10.4103/MJBL.MJBL_93_20.
9. AL-Lami RAH, Al-Hayanni HAS, Shehab ZH. Molecular Investigation of Some Beta-lactamase Genes by PCR and DNA Sequencing Techniques in clinical Escherichia coli. IJS 2022;63(10):4205–4212 https://doi.org/10.24996/ijs.2022.63.10.7.
10. Al-Hayanni HSA. Antimicrobial resistance in Staphylococci and detection of mecA and blaZ genes in Staphylococcus aureus, Staphylococcus sciuri and Staphylococcus epidermidis isolated from fresh beef. Biochem Cell Arch 2021;21(1):1571-1577 https://connectjournals.com/03896.2021.21.1571.
11. Al-Hayanni HAS, El-Shora H. Various Extracts of Some Medicinal Plants as Inhibitors for Beta-lactamase Activity. Baghdad Sci J 2021;18(1):47-48 https://doi.org/10.21123/bsj.2021.18.1.0047
12. Al-Saadi DAA, Al-Mayahi FSA. Antibiogram Susceptibility Patterns of Staphylococcus aureus Harboring of mecA Gene and Prevalence Aminoglycoside Modifying Enzymes (AMEs) Genes in Iraq. IOP Conf Ser: Earth Environ Sci 2021;923: 012049 https://doi.org/10.1088/1755-1315/923/1/012049.
13. Alwash SJ, Aburesha RA. The Differences in Antibiotic Resistance among Several Staphylococcus aureus strains in Iraq. Med Legal Update 2021;21(3):476-485 https://doi.org/10.37506/mlu.v21i3.3034.
14. Du Y, Liu L, Zhang C, Zhang Y. Two residues in Staphylococcus aureus α-hemolysin related to hemolysis and self-assembly. Infect Drug Resist 2018;11:1271-1274 https://doi.org/10.2147/IDR.S167779.
15. Sedarat Z, Taylor-Robinson AW. Consideration of antibacterial agent efficacies in the treatment and prevention of formation of Staphylococcus aureus biofilm. J Microbiol Infect Dis 2019;9(4):167-172 https://doi.org/10.5799/jmid.657903.
16. Jin Z, Jiang Q, Fang B, Sun B. The ArlR-MgrA regulatory cascade regulates PIA-dependent and protein-mediated biofilm formation in Rbf-dependent and Rbf-independent pathways. IJMM 2019; 309(2): 85-96 https://doi.org/10.1016/j.ijmm.2018.12.006
17. Bonar E, Międzobrodzki J, Władyka B. The Staphylococcal Coagulase .Pet-To-Man Travelling Staphylococci 2018;95-102 https://doi.org/10.1016/B978-0-12-813547-1.00007-8.
18. Zhang H, Luan Y, Jing S, Wang Y, Gao Z, Yang P, et. al. Baicalein mediates protection against Staphylococcus aureus-induced pneumonia by inhibiting the coagulase activity of vWbp. Biochem Pharmacol 2020;178: 114024 https://doi.org/10.1016/j.bcp.2020.114024.
19. Clinical and Laboratory Standards Institute (CLSI). “Performance standards for antimicrobial susceptibility testing”; 31st (ed.). Clinical and Laboratory Standard Institute, USA, 2021. https://clsi.org/media/z2uhcbmv/m100ed31_sample.pdf
20. Bauer AW, Kirby WM, Sherris JC, Turck M. “Antibiotic susceptibility testing by a standardized single disk method,”. Am J Clin Pathol 1966;45(4):493-496 https://doi.org/10.1093/ajcp/45.4_ts.493.
21. Shehab ZH, Ahmed ST, and Abdallah N M. Genetic variation of pilB gene in P. aeruginosa”. ATMPH 2020;23(16):SP231615 http://doi.org/10.36295/ASRO.2020.231615.
22. Rocchetti TT, Martins KB, Martins PYF, Oliveira RA, Mondelli AL, Fortaleza CMCB, et.al. Detection of the mecA gene and identification of Staphylococcus directly from blood culture bottles by multiplex polymerase chain reaction. Braz J Infect Dis 2018;22(2):99-105 https://doi.org/10.1016/j.bjid.2018.02.006.
23. Venables WN, Smith DM. The R Core Team. An Introduction to R: Notes on R: A Programming Environment for Data Analysis and Graphics. The Comprehensive R Archive Network. Version 4.3.1, 2023. https://www.scirp.org/reference/referencespapers?referenceid=2294823
24.Yamaguchi T, Ono D, Sato A. Staphylococcal Cassette Chromosome mec (SCCmec) Analysis of MRSA. In: Ji, Y. (eds) Methicillin-Resistant Staphylococcus aureus (MRSA) Protocols. Methods mol biol 2020;2069 https://doi.org/10.1007/978-1-4939-9849-4_4.
25. Scharn CR, Tickler IA, Tenover FC, Goering RV. Characterization of SCCmec Instability in Methicillin-Resistant Staphylococcus aureus Affecting Adjacent Chromosomal Regions, Including the Gene for Staphylococcal Protein A (spa). Antimicrob Agents Chemother 2022;66(4):e0237421 https://doi.org/10.1128/aac.02374-21.
26. Ali AM. Effect of MRSA Irradiation by 632, 532, and 405 nm (Red, Blue, and Green) Diode Lasers on Antibiotic Susceptibility Tests. J Fac Med Bagdad 2017;59(2):191-7 https://doi.org/10.32007/jfacmedbagdad.592136
27. Bastidas CA, Villacrés-Granda I, Navarrete D, Monsalve M, Coral-Almeida M, Cifuentes SG. Antibiotic susceptibility profile and prevalence of mecA and lukS-PV/lukF-PV genes in Staphylococcus aureus isolated from nasal and pharyngeal sources of medical students in Ecuador. Infect Drug Resist 2019;12:2553-2560 https://doi.org/10.2147/IDR.S219358.
28. Ponvelil JJ, Gowda HN, Mysore Ram Raj S. Prevalence of urinary tract infection and sensitivity pattern amongst children less than 3 years of age with fever in a tertiary care hospital in South Karnataka. IJBCP 2020;9(5), 736–742 https://doi.org/10.18203/2319-2003.ijbcp20201749.
29. Salman MK, Ashraf MS, Iftikhar S, Baig MAR. Frequency of nasal carriage of Staphylococcus aureus among health care workers at a Tertiary Care Hospital. Pak J Med Sci 2018;34(5):1181-1184 https://doi.org/10.12669/pjms.345.14588 .
30. Gandhi K, Dhanvijay AK. Antibiotic Susceptibility Pattern of Methicillin Sensitive and Resistant Staphylococcus aureus from Clinical Isolates in a Tertiary Care Hospital at Mathura, Western Uttar Pradesh, J Pure Appl Microbiol 2020;14(1): 455-460 https://doi.org/10.22207/JPAM.14.1.47.
31. Bai Z, Chen M, Lin Q, Ye Y, Fan H, Wen K, et al. Identification of Methicillin-Resistant Staphylococcus aureus From Methicillin-Sensitive Staphylococcus Aureus and Molecular Characterization in Quanzhou, China. Front Cell Dev Biol 2021;9:629681 https://doi.org/10.3389/fcell.2021.629681.
32. Ibrahiem WAM, Rizk DE, Kenawy H, Ebrahim Hassan RH. Prevalence of Vancomycin Resistance among Clinical Isolates of MRSA from Different Governorates in Egypt. Egypt J Med Microbiol 2022;31(4):5-14. https://doi.org/10.21608/EJMM.2022.262673.
33. Khasawneh AI, Himsawi N, Abu-Raideh J, Salameh MA, Al-Tamimi M, Al Haj Mahmoud S, et al. Status of Biofilm-Forming Genes among Jordanian Nasal Carriers of Methicillin-Sensitive and Methicillin-Resistant Staphylococcus aureus. Iran Biomed J 2020;24(6):386-98 https://doi.org/10.29252/ibj.24.6.381
34. Sohail M, Latif Z. Molecular typing of Methicillin Resistance Staphylococcus aureus (MRSA) isolated from device-related infections by SCCmec and PCR-RFLP of coagulase gene. Adv Life Sci 2018;6(1):34-40 http://www.als-journal.com/615-18/.
35. Ndedy MM, Nyasa RB, Esemu SN, Kfusi JA, Keneh NK, Masalla TN, et al. A cross-sectional study on the prevalence and drug susceptibility pattern of methicillin-resistant Staphylococcus aureus isolated from patients in the Buea Health District, Cameroon. Pan Afr Med J 2023;45:28 https://doi.org/10.11604/pamj.2023.45.28.36860.
36. Vieira G, Leal N, Rodrigues A, Chaves C, Rodrigues F, Osório N. MRSA/MSSA causing infections: prevalence of mecA gene. Eur J Public Health 2020;30(2):ckaa040.052 https://doi.org/10.1093/eurpub/ckaa040.052.
37. Valliammai A, Sethupathy S, Priya A, Selvaraj A, Bhaskar JP, Krishnan V et al. 5-Dodecanolide interferes with biofilm formation and reduces the virulence of Methicillin-resistant Staphylococcus aureus (MRSA) through up regulation of agr system. Sci Rep 2019;9(1):13744 https://doi.org/10.1038/s41598-019-50207-y.
38. Bayirli M, Aslantaş Ö, Burçin Ö. Investigation of Toxin Profiles of Methicillin Resistant and Sensitive Staphylococcus aureus Strains Isolated from Various Clinical Specimens. Duzce Med J 2021;23(3):244-251 https://doi.org/10.18678/dtfd.956666
39. Azmi K, Qrei W, Abdeen Z. Screening of genes encoding adhesion factors and biofilm production in methicillin-resistant strains of Staphylococcus aureus isolated from Palestinian patients. BMC Genomics 2019;20:578 https://doi.org/10.1186/s12864-019-5929-1
40. Hezam AM. The phenotypic and genetic characterization of some virulence factors in MRSA isolated from burn patients. J. Phys.: Conf. Ser. 2019; 1294 062061 DOI: 10.1088/1742-6596/1294/6/062061.
Downloads
Published
Issue
Section
License
Copyright (c) 2024 Wed L. Khalil & Huda S. A. Al-Hayanni

This work is licensed under a Creative Commons Attribution 4.0 International License.











 Creative Commons Attribution 4.0 International license..
Creative Commons Attribution 4.0 International license..


