Impact of IDH Mutations on DNA Methylation of Acute Myeloid Leukemia Related Genes: A Review Article
DOI:
https://doi.org/10.32007/jfacmedbagdad.6612175Keywords:
AML; , DNA methylation; , DNA methyltransferases; , IDH;, isocitrate dehydrogenase; , targeted therapy; , TET proteins;Abstract
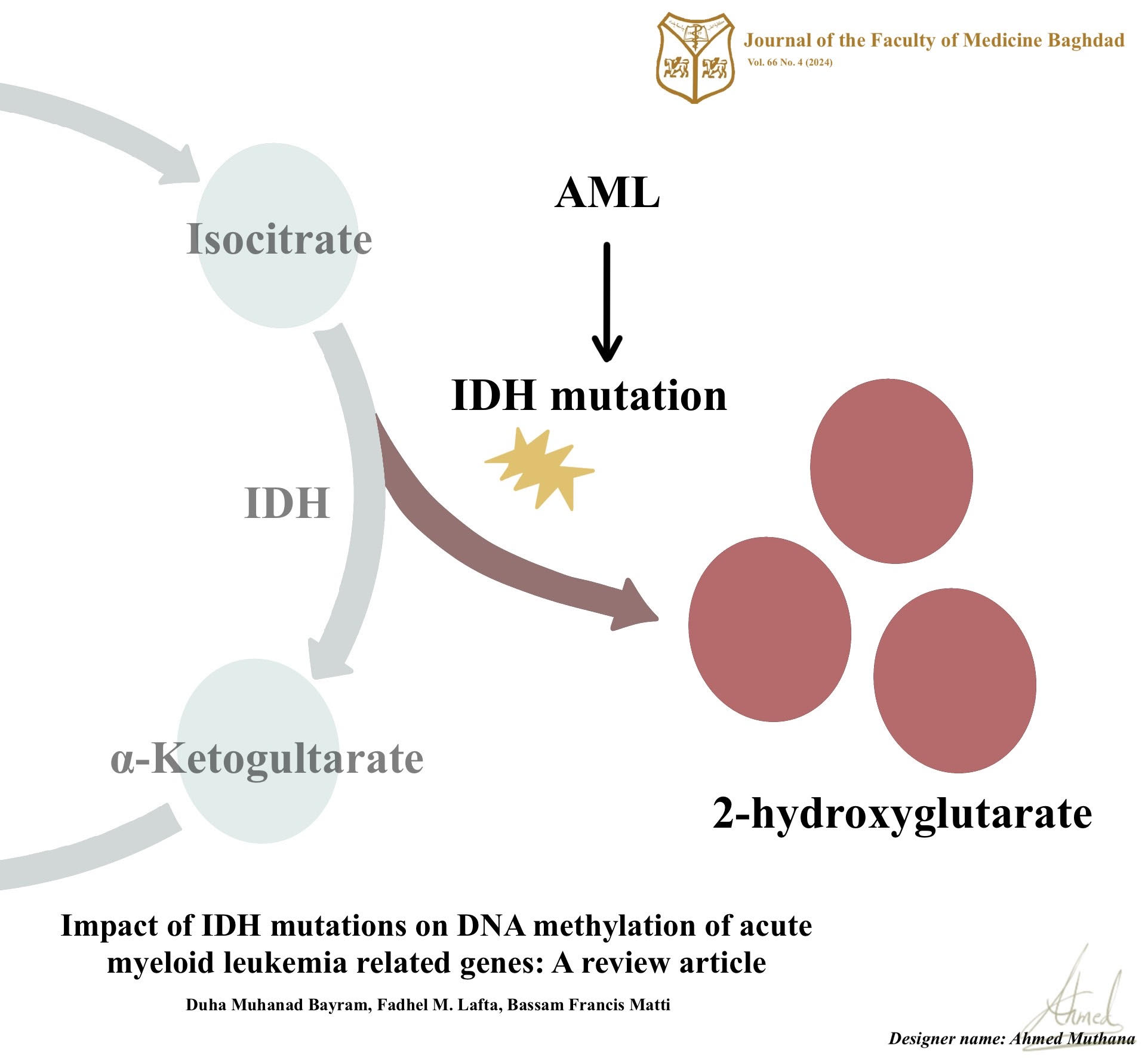
Background: Acute myeloid leukemia is one of the deadliest hematologic malignancies that is marked by genetic alterations, abnormal cellular functions, and proliferation. Mutations in isocitrate dehydrogenase genes, particularly isocitrate dehydrogenase gene 1 and isocitrate dehydrogenase gene 2, have emerged as recurrent genetic abnormalities in acute myeloid leukemia. These mutations lead to abnormal enzymatic activity, accumulating 2-hydroxyglutarate, which disrupts normal cellular processes including DNA methylation.
Objectives: This review article explores recent findings related to the implication of isocitrate dehydrogenase gene mutations on the acute myeloid leukemia epimethylome and provides evidence of the relationship between these mutations and the pathogenesis, prognosis, and treatment of acute myeloid leukemia.
Methods: A comprehensive literature search was conducted to identify relevant studies investigating the impact of isocitrate dehydrogenase mutations on altered DNA methylation patterns of acute myeloid leukemia-related genes. The selected studies were reviewed and analyzed to highlight the significance of their findings.
Results: The review highlights that isocitrate dehydrogenase gene mutations in acute myeloid leukemia are associated with widespread changes in DNA methylation patterns. These alterations primarily affect DNA methylation of acute myeloid leukemia-associated genes, including DNA methyltransferases and ten-eleven translocation proteins. Such epigenetic dysregulation in the DNA methylation modifying genes contributes to global DNA hypermethylation and specific gene hypomethylation leading to abnormal cellular functions and the development of acute myeloid leukemia.
Conclusion: The findings of this review support the significant impact of isocitrate dehydrogenase gene mutations on DNA methylation of acute myeloid leukemia-related genes. Understanding the interplay between isocitrate dehydrogenase gene mutations and DNA methylation dysregulation provides insights into acute myeloid leukemia pathogenicity and may have implications for prognostication and targeted therapies.
Received: June., 2023
Accepted: Sept., 2024
Published: April .2023
Downloads
References
Hamad HM, Shabeeb ZA, Awad MM. Expressions of CD274 (PD-L1) and CD47 rReceptors on the sSurface of bBlast cCells in AML pPatients. IJS. 2020; 63(6), 2373–2387. Available from: https://doi.org/10.24996/ijs.2022.63.6.6
Obeagu EI, Babar Q. Acute Myeloid Leukaemia (AML): The Good, the Bad, and the Ugly. Int J Curr Res Med Sci. 2021;7(7):29-41. Available from: 10.22192/ijcrms.2021.07.07.004
Heimbruch KE, Meyer AE, Agrawal P, Viny AD, Rao S. A cohesive look at leukemogenesis: The cohesin complex and other driving mutations in AML. Neoplasia. 2021;23(3):337-47. DOI: 10.1016/j.neo.2021.01.003
Grimwade D, Ivey A, Huntly BJ. Molecular landscape of acute myeloid leukemia in younger adults and its clinical relevance. Blood, The Journal of the American Society of Hematology. 2016;127(1):29-41. DOI: 10.1182/blood-2015-07-604496
Stelmach P, Trumpp A. Leukemic stem cells and therapy resistance in acute myeloid leukemia. Haematologica. 2023;108(2):353. https://doi.org/10.3324/haematol.2022.280800
Al-Mudallel SS, Dhahi MA. Detection of FLT3-ITDMutation in Twenty ChildwithAcuteMyeloidLeukemia in One Iraqi Teaching Hospital. Journal of the Faculty of Medicine Baghdad. 2012;54(2):138-41. Available from: https://doi.org/10.24996/ijs.2022.63.6.6
Zhang N, Chen Y, Lou S, Shen Y, Deng J. A six-gene-based prognostic model predicts complete remission and overall survival in childhood acute myeloid leukemia. Onco Targets Ther. 2019;12:6591.https://doi.org/10.2147/OTT.S218928
Elgarten CW, Aplenc R. Pediatric acute myeloid leukemia: updates on biology, risk stratification, and therapy. Curr Opin Pediatr. 2020;32(1):57-66. Available from: https://doi.org/10.1097/MOP.0000000000000855
AL-Khalissi KA. Experience with treatment of fifty eight Iraqi patients with acute myeloid leukemia. Journal of the Faculty of Medicine Baghdad. 2013;55(4):290-5. Available from: https://doi.org/10.32007/jfacmedbagdad.554566 .
Pelcovits A, Niroula R. Acute myeloid leukemia: a review. Rhode Island Medical Journal. 2020;103(3):38-40. 2020-04-38-hem-onc-pelcovits.pdf (rimed.org)
Walker CJ, Mrózek K, Ozer HG, Nicolet D, Kohlschmidt J, Papaioannou D, et al. Gene expression signature predicts relapse in adult patients with cytogenetically normal acute myeloid leukemia. Blood advances. 2021;5(5):1474-82. https://doi.org/10.1182/bloodadvances.2020003727
Herold T, Rothenberg-Thurley M, Grunwald VV, Janke H, Goerlich D, Sauerland MC, et al. Validation and refinement of the revised 2017 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia. 2020;34(12):3161-72. https://doi.org/10.1038/s41375-020-0806-0
Rausch C, Rothenberg-Thurley M, Dufour A, Schneider S, Gittinger H, Sauerland C, et al. Validation and refinement of the 2022 European LeukemiaNet genetic risk stratification of acute myeloid leukemia. Leukemia. 2023:1-11. https:/doi.org/10.1038/s41375-023-01884-2
Mohammed SK, AL-Faisal AHM. Study of Chromosomal Aberrations and Micronucleus Formation in Some Iraqi Patients infected with Acute Myeloid Leukemia (AML). Iraqi journal of biotechnology. 2014;13(1). Available from: https://jige.uobaghdad.edu.iq/index.php/IJB/article/view/264
Ferrer A, Stephens ZD, Kocher JP. Experimental and Computational Approaches to Measure Telomere Length: Recent Advances and Future Directions. Current Hematologic Malignancy Reports. 2023 Dec;18(6):284-91. https://doi.org/10.1007/s11899-023-00717-4
Celton M, Forest A, Gosse G, Lemieux S, Hebert J, Sauvageau G, et al. Epigenetic regulation of GATA2 and its impact on normal karyotype acute myeloid leukemia. Leukemia. 2014;28(8):1617-26. Available from: https://doi.org/10.1038/leu.2014.67
Sandoval J, Heyn H, Moran S, Serra-Musach J, Pujana MA, Bibikova M, et al. Validation of a DNA methylation microarray for 450,000 CpG sites in the human genome. Epigenetics. 2011;6(6):692-702. https://doi.org/10.4161/epi.6.6.16196
Cheng Y, He C, Wang M, Ma X, Mo F, Yang S, et al. Targeting epigenetic regulators for cancer therapy: mechanisms and advances in clinical trials. Signal transduction and targeted therapy. 2019;4(1):62. Available from: https://doi.org/10.3390/ijms22052305.
Lu Y, Chan Y-T, Tan H-Y, Li S, Wang N, Feng Y. Epigenetic regulation in human cancer: the potential role of epi-drug in cancer therapy. Mol Cancer. 2020;19(1):1-16. Available from: https://doi.org/10.1186/s12943-020-01197-3
Pulikkottil AJ, Bamezai S, Ammer T, Mohr F, Feder K, Vegi NM, et al. TET3 promotes AML growth and epigenetically regulates glucose metabolism and leukemic stem cell associated pathways. Leukemia. 2022;36(2):416-25. https://doi.org/10.1038/s41375-021-01390-3
Zhang L, Lu Q, Chang C. Epigenetics in health and disease. Epigenetics in allergy and autoimmunity. 2020:3-55. https:doi.org/ 10.1007/978-981-15-3449-2_1
Ren W, Fan H, Grimm SA, Guo Y, Kim JJ, Yin J, et al. Direct readout of heterochromatic H3K9me3 regulates DNMT1-mediated maintenance DNA methylation. Proceedings of the National Academy of Sciences. 2020;117(31):18439-47. https:doi.org/10.1073/pnas.2009316117
Heredia-Mendez AJ, Sánchez-Sánchez G, López-Camarillo C. Reprogramming of the Genome-Wide DNA Methylation Landscape in Three-Dimensional Cancer Cell Cultures. Cancers (Basel). 2023;15(7):1991. https:doi.org/10.3390/cancers15071991
Gao L, Emperle M, Guo Y, Grimm SA, Ren W, Adam S, et al. Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nature communications. 2020;11(1):1-14.https://doi.org/10.1038/s41467-020-17109-4
Shekhawat J, Gauba K, Gupta S, Choudhury B, Purohit P, Sharma P, et al. Ten–eleven translocase: key regulator of the methylation landscape in cancer. J Cancer Res Clin Oncol. 2021;147(7):1869-79. https:doi.org/10.1007/s00432-021-03641-3
Zeng Y. Regulation of de novo and Maintenance DNA Methylation by DNMT3A and DNMT3B. 2023. https://digitalcommons.library.tmc.edu/utgsbs_dissertations/1264/
Joshi K, Liu S, Breslin SJ P, Zhang J. Mechanisms that regulate the activities of TET proteins. Cell Mol Life Sci. 2022;79(7):363. https://doi.org/0.1007/s00018-022-04396-x
Jiang S. Tet2 at the interface between cancer and immunity. Communications biology. 2020;3(1):667. https://doi.org/10.1038/s42003-020-0139.
Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324(5930):1076-80. https://doi.org10.1126/science.1164097
Nicholson TB, Veland N, Chen T. Writers, readers, and erasers of epigenetic marks. Epigenetic Cancer Therapy: Elsevier; 2015. p. 31-66. https://doi.org/0.1016/B978-0-12-800206-3.00003-3
Raineri S, Mellor J. IDH1: linking metabolism and epigenetics. Frontiers in genetics. 2018:493. https://doi.org/10.3389/fgene.2018.00493
Nikolaev A, Fiveash JB, Yang ES. Combined targeting of mutant p53 and Jumonji family histone demethylase augments therapeutic efficacy of radiation in H3K27M DIPG. Int J Mol Sci. 2020;21(2):490. https:.org/10.3390/ijms21020490
Lee C-J, Ahn H, Jeong D, Pak M, Moon JH, Kim S. Impact of mutations in DNA methylation modification genes on genome-wide methylation landscapes and downstream gene activations in pan-cancer. BMC Med Genomics. 2020;13:1-14. Available from: https://doi.org/10.1186/s12920-020-0659-4
Pelosi E, Castelli G, Testa U. Isocitrate dehydrogenase mutations in human cancers: physiopathologic mechanisms and therapeutic targeting. Journal of Exploratory Research in Pharmacology. 2016;1(1):20-34. DOI: 10.14218/JERP.2016.00019
Senhaji N, Squalli Houssaini A, Lamrabet S, Louati S, Bennis S. Molecular and circulating biomarkers in patients with glioblastoma. Int J Mol Sci. 2022;23(13):7474. Available from: https://doi.org/10.3390/ijms23137474
Willander K, Falk IJ, Chaireti R, Paul E, Hermansson M, Gréen H, et al. Mutations in the isocitrate dehydrogenase 2 gene and IDH1 SNP 105C> T have a prognostic value in acute myeloid leukemia. Biomarker research. 2014;2(1):1-9. Available from: https://doi.org/10.1186/2050-7771-2-18
Alkhatabi H, Bin Saddeq HA, Alyamani L, Shinawi T, Yasin EB, Alserihi R, et al. Investigation of Isocitrate Dehydrogenase 1 and 2 Mutations in Acute Leukemia Patients in Saudi Arabia. Genes. 2021;12(12):1963. Available from: https://doi.org/10.3390/genes12121963.
Cumbo C, Minervini CF, Orsini P, Anelli L, Zagaria A, Minervini A, et al. Nanopore targeted sequencing for rapid gene mutations detection in acute myeloid leukemia. Genes. 2019;10(12):1026. PMID: 31835432
Rausch C, Rothenberg-Thurley M, Buerger SA, Tschuri S, Dufour A, Neusser M, et al. Double drop-off droplet digital pcr: A novel, versatile tool for mutation screening and residual disease monitoring in acute myeloid leukemia using cellular or cell-free DNA. The Journal of Molecular Diagnostics. 2021;23(8):975-85. https:doi.org/10.1016/j.jmoldx.2021.05.001
Beck RC, Kim AS, Goswami RS, Weinberg OK, Yeung CC, Ewalt MD. Molecular/Cytogenetic Education for Hematopathology Fellows: A Recommended Curriculum From the Society for Hematopathology and the Association for Molecular Pathology. Am J Clin Pathol. 2020;154(2):149-77. Available from: https://doi.org/10.1093/ajcp/aqaa038.
Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739-44. Available from: https://doi.org/10.1038/nature08617
Heiblig M. Les mutations d'IDH2 dans la leucémie aigue myéloïde confèrent une sensibilité spécifique à l'inhibition du métabolisme des acides aminés branchés (BCAA): Université Paris-Saclay; 2022. Available from: https://theses.hal.science/tel-04108390/document
DiNardo CD, Propert KJ, Loren AW, Paietta E, Sun Z, Levine RL, et al. Serum 2-hydroxyglutarate levels predict isocitrate dehydrogenase mutations and clinical outcome in acute myeloid leukemia. Blood, The Journal of the American Society of Hematology. 2013;121(24):4917-24. https:doi.org/10.1182/blood-2013-03-493197
Pang H, Jia W, Hu Z. Emerging applications of metabolomics in clinical pharmacology. Clin Pharmacol Ther. 2019;106(3):544-56. https:doi.org/10.1002/cpt.1538
Losman J-A, Looper RE, Koivunen P, Lee S, Schneider RK, McMahon C, et al. (R)-2-hydroxyglutarate is sufficient to promote leukemogenesis and its effects are reversible. Science. 2013;339(6127):1621-5. https://doi.org/10.1126/science.1231677
Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553-67. https:/doi.org/10.1016/j.ccr.2010.11.015
Zhou H, Zhang Q, Huang W, He C, Zhou C, Zhou J, et al. Epigenetic silencing of ZCCHC10 by the lncRNA SNHG1 promotes progression and venetoclax resistance of acute myeloid leukemia. Int J Oncol. 2023;62(5):1-11. DOI: 10.3892/ijo.2023.5512
Angelakas A, Lamarca A, Hubner RA, McNamara MG, Valle JW. Ivosidenib: An investigational drug for the treatment of biliary tract cancers. Expert Opinion on Investigational Drugs. 2021;30(4):301-7. Available from: https://doi.org/10.1080/13543784.2021.1900115.
Sun Y, Chen B-R, Deshpande A. Epigenetic regulators in the development, maintenance, and therapeutic targeting of acute myeloid leukemia. Front Oncol. 2018;8:41. Available from: https://doi.org/10.3389/fonc.2018.00041
Davis AR, Canady BC, Aggarwal N, Bailey NG. Clinicopathologic Features of IDH2 R172–Mutated Myeloid Neoplasms. Am J Clin Pathol. 2023:aqad019. DOI: 10.1093/ajcp/aqad019
Bill M, Jentzsch M, Bischof L, Kohlschmidt J, Grimm J, Schmalbrock LK, et al. Impact of IDH1 and IDH2 mutation detection at diagnosis and in remission in patients with AML receiving allogeneic transplantation. Blood Advances. 2023;7(3):436-44. Available from: https://doi.org/10.1002/onco.13656.
Abou Dalle I, DiNardo CD. The role of enasidenib in the treatment of mutant IDH2 acute myeloid leukemia. Therapeutic advances in hematology. 2018;9(7):163-73. Available from: https://doi.org/10.1177/2040620718777467.
Beat A. Study Offers Insights on Prognostic Significance of IDH Mutations Across Age Groups in AML. The Oncologist. 2020;26:S13–S4. Available from: https://doi.org/10.1002/onco.13656.
Myers RA, Wirth S, Williams S, Kiel PJ. Enasidenib: an oral IDH2 inhibitor for the treatment of acute myeloid leukemia. Journal of the advanced practitioner in oncology. 2018;9(4):435. PMID: 30719396
Aiman W, Ali MA, Basit MA, Omar Z, Suleman M, Hassan M, et al. Efficacy and Tolerability of Isocitrate Dehydrogenase Inhibitors in Patients with Acute Myeloid Leukemia: A Systematic Review of Clinical Trials. Leuk Res. 2023:107077. Available from: https://doi.org/10.1016/j.leukres.2023.107077.
McDevitt MA, editor Clinical applications of epigenetic markers and epigenetic profiling in myeloid malignancies. Semin Oncol; 2012: Elsevier.
Montalban-Bravo G, DiNardo CD. The role of IDH mutations in acute myeloid leukemia. Future Oncology. 2018;14(10):979-93. DOI: 10.2217/fon-2017-0523
Liu X, Gong Y. Isocitrate dehydrogenase inhibitors in acute myeloid leukemia. Biomarker research. 2019;7(1):1-8. DOI: https://doi.org/10.1186/s40364-019-0173-z
Wang J, Tomlinson B, Lazarus HM. Update on Small Molecule Targeted Therapies for Acute Myeloid Leukemia. Curr Treat Options Oncol. 2023:1-32. DOI: 10.1007/s11864-023-01090-3
Venugopal S, Watts JM. Olutasidenib: from bench to bedside. Blood Advances. 2023:bloodadvances. 2023; 009854. https:doi.org/10.1182/bloodadvances.2023009854
Leotta S, Condorelli A, Sciortino R, Milone GA, Bellofiore C, Garibaldi B, et al. Prevention and treatment of acute myeloid leukemia relapse after hematopoietic stem cell transplantation: the state of the art and future perspectives. Journal of Clinical Medicine. 2022;11(1):253. https://doi.org/10.3390/jcm11010253
Testa U, Pelosi E, Castelli G. Cancer stem cell targeted therapies. Ann Ist Super Sanita. 2020;56(3):336-50. http://doi.org/10.4415/ANN_20_03_12
Thol F, Ganser A. Treatment of relapsed acute myeloid leukemia. Curr Treat Options Oncol. 2020;21(8):1-11. https://doi.org/10.1007/s11864-020-00765-5
Guinn B, Schuler PJ, Schrezenmeier H, Hofmann S, Weiss J, Bulach C, et al. A combination of the immunotherapeutic drug anti-programmed death 1 with lenalidomide enhances specific T cell immune responses against acute myeloid leukemia cells. Int J Mol Sci. 2023;24(11):9285. DOI: 10.3390/ijms24119285
AL‐KZAYER, Lika'a Fasih Y., et al. Analysis of KRAS and NRAS gene mutations in Arab Asian children with acute leukemia: high frequency of RAS mutations in acute lymphoblastic leukemia. Pediatric blood & cancer. 2015, 62.12: 2157-2161.
Bashi MA, Ad'hiah AH. Interleukin-37 gene expression is down-regulated in patients with acute myeloid leukemia and shown to be affected by CD14 and HLA-DR immunophenotypes. Cytokine. 2023 Nov;171:156368.https://doi.org/10.1016/j.cyto.2023.156368
Bashi MA, Ad'hiah AH. Molecular landscape of the interleukin-40 encoding gene, C17orf99, in patients with acute myeloid leukemia. Gene. 2024 Apr 30;904:148214. https:doi.org/10.1016/j.gene.2024.148214.
Downloads
Additional Files
Published
Issue
Section
License
Copyright (c) 2024 Duha M. Bayram, Fadhel M. Lafta, Bassam F. Matti

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.











 Creative Commons Attribution 4.0 International license..
Creative Commons Attribution 4.0 International license..


