Detection of Class 1 Integron among Klebsiella pneumoniae Clinical Isolates in Baghdad Hospitals
DOI:
https://doi.org/10.32007/jfacmedbaghdad3161Keywords:
Clinical isolates; , Class 1 integron; , Klebsiella pneumoniae; , MDR; , Nosocomial infections; , PCR.Abstract
Background: Extensively drug-resistant Klebsiella pneumoniae (K. pneumoniae) is a significant problem currently due to the increasing prevalence of this pathogen.
Objectives: To underscore the growing threat of antibiotic-resistant K. pneumoniae by investigating the prevalence of class 1 integrons among selected multidrug-resistant (MDR) clinical isolates, with the goal of emphasizing the need for strengthened surveillance and targeted antimicrobial strategies.
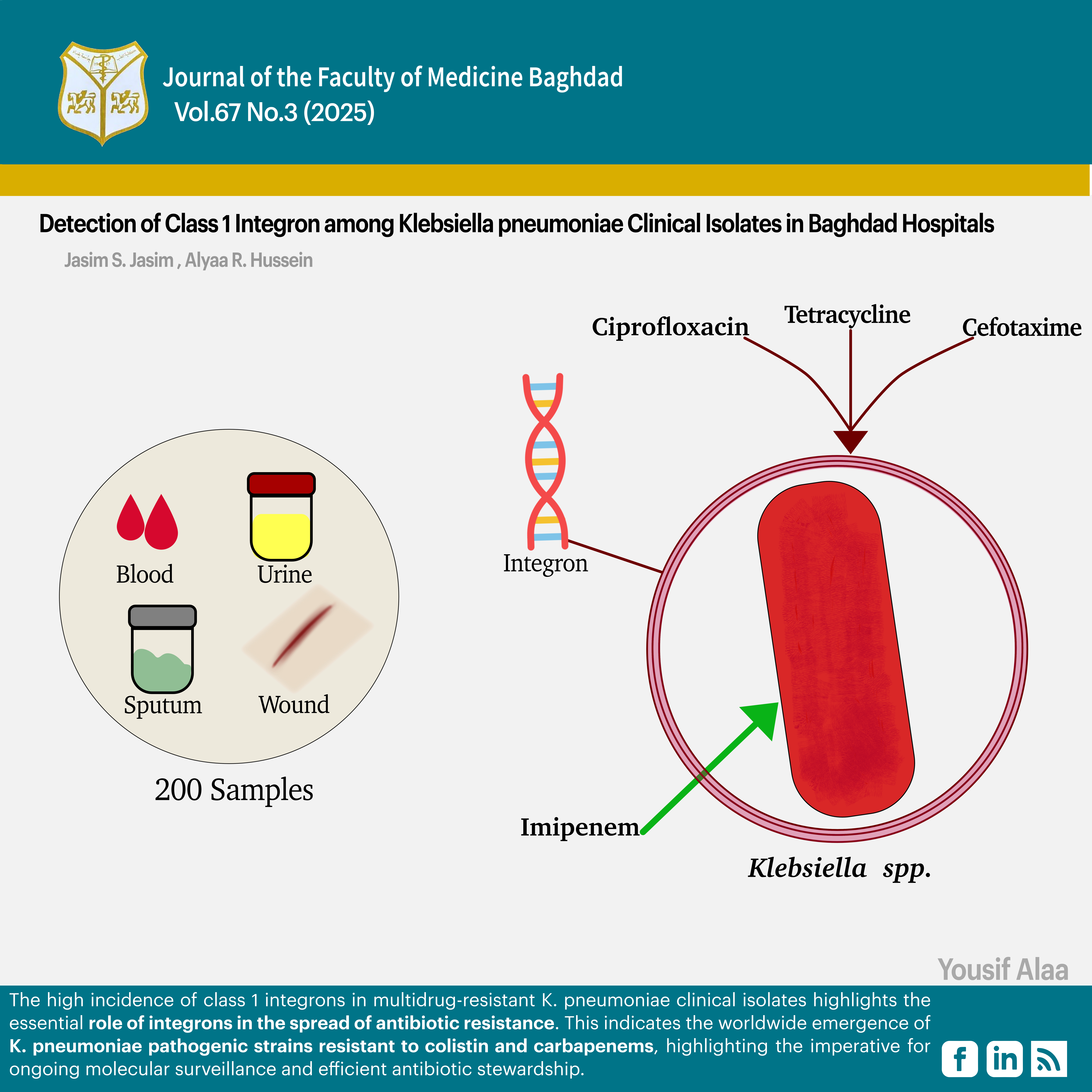
Methods: Seventy-four K. pneumoniae isolates have been identified out of 200 clinical samples from different clinical sources (urine, burns, blood, sputum, wounds). Isolates were obtained from November 2024 to March 2025, in Al-Kadhimiya Teaching Hospital and the Laboratory at College of Science Microbiology Department, University of Baghdad. The isolates were first detected by biochemical testing, chromogenic agar and later confirmed using the VITEK 2 method. Antimicrobial susceptibility was evaluated utilizing the disc diffusion technique (Kirby-Bauer method). The presence of the class 1 integron gene was confirmed via conventional PCR.
Results: A total of 200 clinical samples were collected from the 74 K. pneumoniae isolates; 51 (68.9%) exhibited multidrug resistance. Resistance to cefotaxime was observed in 86%, followed by tetracycline (80%), while imipenem exhibited the highest sensitivity at 84%. Class 1 integron gene was identified in 80% of the 10 selected multidrug-resistant isolates.
Conclusion: The high incidence of class 1 integrons in multidrug-resistant K. pneumoniae clinical isolates highlights the essential role of integrons in the spread of antibiotic resistance. This indicates the worldwide emergence of K. pneumoniae pathogenic strains resistant to colistin and carbapenems, highlighting the imperative for ongoing molecular surveillance and efficient antibiotic stewardship.
Received: May 2025
Revised: Aug. 2025
Accepted: Sept. 2025
Published Online: Sept. 2025
Published: Oct. 2025
References
1. Bai R, Guo J. Interactions and implications of Klebsiella pneumoniae with human immune responses and metabolic pathways: a comprehensive review. Infect Drug Resist. 2024;17:449–62. https://doi.org/10.2147/IDR.S451013
2. Chakraverty R, Kundu AK. Hospital-Acquired Infections in Intensive Care Unit and their Management: The Indian Perspective. Springer; 2025. https://link.springer.com/book/10.1007/978-981-96-0018-2
3. Razzaq T, Ali Y, Khan S, Nadeem Z, Batool A, Ibraheem M, et al. Antibacterial drug exposure and risk of carbapenem-resistant Klebsiella pneumoniae: a review. Zenodo. 2025;3(2):3007–1593. https://doi.org/10.5281/zenodo.15279758
4. Albermani M, Salman WA, Hamad GK, Hadi EA. Infection control measures to reduce hospital infection rates in the medical city burn center. J Fac Med Baghdad. 2019;60(4):191–4. https://doi.org/10.32007/med.1936/jfacmedbagdad.v60i4.3
5. Russo A, Fusco P, Morrone HL, Trecarichi EM, Torti C. New advances in management and treatment of multidrug-resistant Klebsiella pneumoniae. Expert Rev Anti Infect Ther. 2023;21(1):41–55. https://doi.org/10.1080/14787210.2023.2151435
6. Gidla VVS, Anjaneyulu K, Bhanu RV, Dinesh G. Integrons: a mobile genetic element of concern in antimicrobial resistance gene transfer: an overview. Indian J Vet Sci Biotechnol. 2023;19(3):1–5. https://doi.org/10.48165/ijvsbt.19.3.01
7. Tao S, Chen H, Li N, Liang W. The application of the CRISPR-Cas system in antibiotic resistance. Infect Drug Resist. 2022;15:4155–68. https://doi.org/10.2147/IDR.S370869
8. Wang L, Zhu M, Yan C, Zhang Y, He X, Wu L, et al. Class 1 integrons and multiple mobile genetic elements in clinical isolates of the Klebsiella pneumoniae complex from a tertiary hospital in eastern China. Front Microbiol. 2023;14:985102. https://doi.org/10.3389/fmicb.2023.985102
9. Taha RQ, Mansoor RJ, Kareem SA, Mohsein OA. Unraveling the genomic landscape of multidrug-resistant Klebsiella pneumoniae: a focus on resistance islands. Caspian J Med Nat Sci. 2025;6(2). https://doi.org/10.17605/cajmns.v6i2.2754
10. Omar FH, Ibrahim AH. The prevalence of integron class I and II among multidrug-resistant Klebsiella pneumoniae. Iraqi J Agric Sci. 2023;54(3):619–29. https://doi.org/10.36103/ijas.v54i3.1775
11. Alara JA, Alara OR. An overview of the global alarming increase of multiple drug resistant: a major challenge in clinical diagnosis. Infect Disord Drug Targets. 2024;24(3). https://doi.org/10.2174/1871526523666230725103902
12. Al-Ruobayiee MR, Ibrahim AH. The relationship between OqxAB efflux pump and drug resistance in Klebsiella pneumoniae isolated from clinical sources. Al-Rafidain J Med Sci. 2023;5(1):S106–12. https://doi.org/10.54133/ajms.v5i1S.309
13. Alansary IMM, Al-Saryi NA. Emergence of hypervirulent Klebsiella pneumoniae isolates from some Iraqi hospitals. Rev Res Med Microbiol. 2024;35(2):88–96. https://doi.org/10.1097/MRM.0000000000000354
14. CLSI. Performance standards for antimicrobial susceptibility testing. CLSI supplement M100. Clinical and Laboratory Standards Institute; 2024. https://clsi.org/shop/standards/m100
15. Abdel-Hamid RM, El-Mahallawy HA, Allam RM, Zafer MM, Elswify M. Changing patterns of bacterial profile and antimicrobial resistance in high-risk patients during the COVID-19 pandemic at a tertiary oncology hospital. Arch Microbiol. 2024;206(6). https://doi.org/10.1007/S00203-024-03965-X
16. Firoozeh F, Mahluji Z, Khorshidi A, Zibaei M. Molecular characterization of class 1, 2 and 3 integrons in clinical multidrug-resistant Klebsiella pneumoniae isolates. Antimicrob Resist Infect Control. 2019;8(1). https://doi.org/10.1186/s13756-019-0509-3
17. Mustafa SS, Batool R, Kamran M, Javed H, Jamil N. Evaluating the role of wastewaters as reservoirs of antibiotic-resistant ESKAPEE bacteria using phenotypic and molecular methods. Infect Drug Resist. 2022;15:5715–28. https://doi.org/10.2147/IDR.S368886
18. Al-Rubaye DS, Hamza HM, Abdulrahman TR. Genotyping of Klebsiella spp. isolated from different clinical sources. Iraqi J Sci. 2016;57(3B):1937–51. https://doi.org/10.24996/ijs.2016.57.3B.21
19. Abd Al-Hamed ZA, Abd Al-Mayahi FS. Antibiogram of Klebsiella pneumoniae isolated from clinical and environmental samples in Al-Diwaniyah hospitals. Al-Qadisiyah J Pure Sci. 2021;26(2):44–54. https://doi.org/10.29350/qjps.2021.26.2.1296
20. Mahmood MK, Farhan SD, Ahmed BA, Nori W. Risk factors analysis of urosepsis following retrograde intrarenal surgery: a retrospective study. Al-Rafidain J Med Sci. 2025;8(1):48–55. https://doi.org/10.54133/ajms.v8i1.1643
21. Turki Monawer A. Molecular analysis of Klebsiella pneumoniae isolates collected from sputum samples in Duhok, Iraq. Al-Nahrain J Sci. 2025;28(1):91–5. https://doi.org/10.22401/ANJS.28.1.10
22. Jain C, Nikita N. Evaluation of chromogenic agar media for isolation, identification and direct antibiotic susceptibility testing of uropathogens. Int J Pharm Res Allied Sci. 2023;12(2):7–12. https://doi.org/10.51847/Kd4vp42v9B
23. Kherd A, Saaban A, Adyee, Ibrahim Fadhel IE. Wang-ball polynomials for the numerical solution of singular ordinary differential equations. Iraqi J Sci. 2021;62(3):941–9. https://doi.org/10.24996/ijs.2021.62.3.24
24. Ibraheim HK, Madhi KS, Jasim AS, Gharban HAJ. Molecular identification of Klebsiella species from pneumonic goats, Iraq. Open Vet J. 2024;14(11):2980–8. https://doi.org/10.5455/OVJ.2024.v14.i11.26
25. Hmood AH, Alammar MHM. Genotypic and phenotypic surveillance of multidrug resistance of hypermucoid K. pneumoniae among clinical isolates in Al-Najaf City, Iraq. Al-Kufa Univ J Biol. 2023;15(3):122–8. https://doi.org/10.36320/ajb/v15.i3.14615
26. Chhabra P, Rouhani S, Browne H, Peñataro Yori P, Siguas Salas M, Paredes Olortegui M, et al. Homotypic and heterotypic protection and risk of reinfection following natural norovirus infection in a highly endemic setting. Clin Infect Dis. 2021;72(2):222–9. https://doi.org/10.1093/cid/ciaa019
27. García-Fernández S, Simner PJ, Thomson G, Faron M, Bosboom R, van Griethuijsen A, et al. Rapid identification from rectal swabs of the clinically most relevant carbapenemase genes from gram-negative bacteria using the BD MAX Check-Points CPO assay. Diagn Microbiol Infect Dis. 2022;102(1):115554. https://doi.org/10.1016/j.diagmicrobio.2021.115554
28. Jabar ZA, Auhim HS, Hussein AR. Molecular detection of fimH and mrkD genes of strong biofilm producers and MDR Klebsiella pneumoniae. Int J Health Sci. 2022;6(S4):9225–35. https://doi.org/10.53730/ijhs.v6ns4.11954
29. Chowdhury MSR, Hossain H, Rahman MN, Hossain MA, Saha O, Zinnah MA, et al. Emergence of highly virulent multidrug- and extensively drug-resistant Escherichia coli and Klebsiella pneumoniae in buffalo subclinical mastitis cases. Sci Rep. 2025;15:11704. https://doi.org/10.1038/s41598-025-95914-x
30. Al-Marzooq F, Mohd Yusof MY, Tay ST. Molecular analysis of antibiotic resistance determinants and plasmids in Malaysian isolates of multidrug-resistant Klebsiella pneumoniae. PLoS One. 2015;10(7):e0133654. https://doi.org/10.1371/journal.pone.0133654
31. Almidhatee HRM, Alkufaishi ZHJ, Al-Saffar AKH, Alhamawandi JAA. Detection of molecular resistance mechanisms in Klebsiella pneumoniae through carbapenem enzymatic genes. J Angiother. 2024;8(4). https://doi.org/10.25163/angiotherapy.849602
32. Fouad NA, Totl KJ. Antibiotic resistance in Klebsiella pneumoniae and its impact in mixed type isolates. Iraqi Journal of Medical Sciences. 2024;22(2):369–376. doi:10.22578/IJMS.22.2.22 https://iraqijms.net/index.php/jms/article/view/1059
33. Al-Qaysi AMK, Ahmed MM, Habeeb WH, Al-Meani SAL, Janaby MS AL, Alalwani AK, et al. Genetic variants of multidrug-resistant Klebsiella pneumoniae isolated from Al-Ramadi Teaching Hospital, Iraq. Open Microbiol J. 2024;18(1). https://doi.org/10.2174/0118742858298979240628070603
34. Mohammed MJ, Mahmood SS. The prevalence of pks genotoxin among Klebsiella pneumoniae isolated from different clinical samples in Baghdad, Iraq. Iraqi J Sci. 2024;65(7):3716–24. https://doi.org/10.24996/ijs.2024.65.7.13
35. Jazayeri Moghadas A, Kalantari F, Sarfi M, Shahhoseini S, Mirkalantari S. Evaluation of virulence factors and antibiotic resistance patterns in clinical urine isolates of Klebsiella pneumoniae in Semnan, Iran. Jundishapur J Microbiol. 2018;11(7). https://doi.org/10.5812/jjm.63637
36. Fadare FT, Fadare TO, Okoh AI. Prevalence and molecular characterization of integrons and associated gene cassettes in Klebsiella pneumoniae and K. oxytoca recovered from diverse environmental matrices. Sci Rep. 2023;13(1). https://doi.org/10.1038/s41598-023-41591-7
37. Sheikh AF, Abdi M, Farshadzadeh Z. Molecular detection of Class 1, 2, and 3 integrons in hypervirulent and classic Klebsiella pneumoniae isolates: a cross-sectional study. Health Sci Rep. 2024;7(5):e1962. https://doi.org/10.1002/hsr2.1962
38. Hetta HF, Alanazi FE, Sayed Ali MA, Alatawi AD, Aljohani HM, Ahmed R, et al. Hypervirulent Klebsiella pneumoniae: Insights into Virulence, Antibiotic Resistance, and Fight Strategies Against aSuperbug. Pharmaceuticals (Basel). 2025;18(5):724. https://doi.org/10.3390/ph18050724
Downloads
Published
Issue
Section
Categories
License
Copyright (c) 2025 Jasim S. Jasim, Alyaa R. Hussein

This work is licensed under a Creative Commons Attribution 4.0 International License.










 Creative Commons Attribution 4.0 International license..
Creative Commons Attribution 4.0 International license..


